
Where X_is are the original variables, and Beta_is are the corresponding weights or so called coefficients. components_ attribute.Įach principal component is a linear combination of the original variables: For an intuition about this, think about a vector and its negative in 3-D space - both are essentially representing the same direction in space.Įdit: as others have commented, you may get same values from.
#Add pca column back to data code
In fact, if you run the PCA code again, you might get the PCA dimensions with the signs inverted. Only the relative signs of features forming the PCA dimension are important. IMPORTANT: As a side comment, note the PCA sign does not affect its interpretation since the sign does not affect the variance contained in each component.
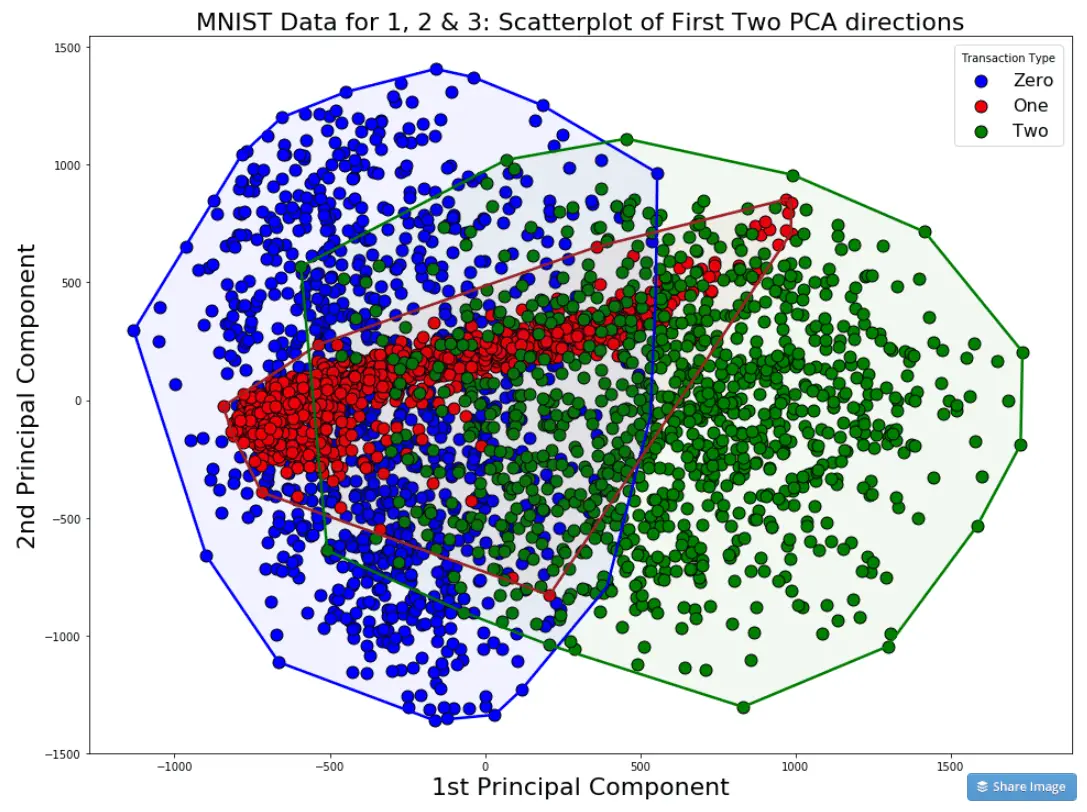
Sepal length (cm) sepal width (cm) petal length (cm) petal width (cm) Print(pd.DataFrame(pca.components_,columns=data_lumns,index = )) # Dump components relations with features: Note: each coefficient represents the correlation between a particular pair of component and feature import pandas as pdĭf = pd.DataFrame(iris.data, columns=iris.feature_names)ĭata_scaled = pd.DataFrame(preprocessing.scale(df),columns = df.columns)
As described in the documentation, pca.components_ outputs an array of, so to get how components are linearly related with the different features you have to: This information is included in the pca attribute: components_.


 0 kommentar(er)
0 kommentar(er)
